Cancers Free Full Text Exploring Spatial Heterogeneity Of Immune

Cancers Free Full Text Exploring Spatial Heterogeneity Of Immune The paper "exploring spatial heterogeneity of immune cells in nasopharyngeal cancer" by sobti and colleagues describes the profiling of 43 proteins in various immune cells from ffpe slides from previously analyzed nasopharingeal cancer (npc) samples. the study is interesting but it could be made more robust and convincing. Feature papers represent the most advanced research with significant potential for high impact in the field. a feature paper should be a substantial original article that involves several techniques or approaches, provides an outlook for future research directions and describes possible research applications.
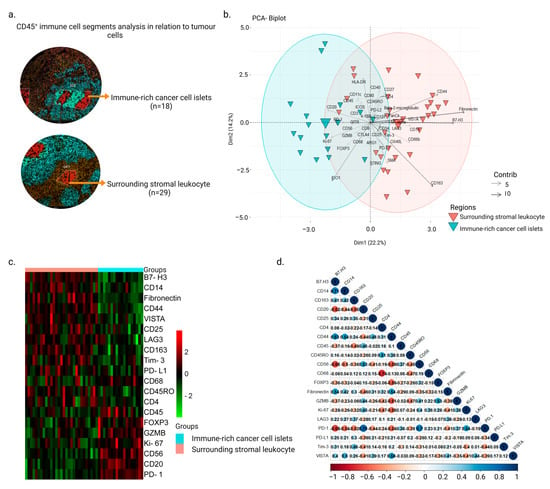
Cancers Free Full Text Exploring Spatial Heterogeneity Of Immune Abstract. nasopharyngeal cancer (npc) is a malignant tumor. in a recent publication, we described the presence and distribution of cd8 t cells in npc and used the information to identify 'inflamed', 'immune excluded', and 'desert' immune phenotypes, where 'inflamed' and 'immune excluded' npcs were correlated with cd8 t cell infiltration and. Simple summary. nasopharyngeal cancer (npc) is a malignant tumor of the upper pharynx. in this study, we used multiplex tissue analysis combined with digital spatial profiling to map the immune heterogeneity of npc. forty seven specified regions of interest (rois) with 49 target proteins were profiled across 30 cases of npc for quantitative. Background nasopharyngeal cancer (npc) is a squamous cell carcinoma of the upper pharynx, strongly associated with epstein barr virus (ebv), and with varying incidences in the world. we have previously investigated the presence and distribution of cd8 t cells in npc and found that an ‘ inflamed profile ’ , with cd8 t cells in and around cancer cell areas, was associated with better. Spatially resolved multi omics revolutionizes cancer therapy by decoding the cellular and molecular heterogeneity of the tumor microenvironment through spatial coordinates. this commentary discusses the roles of spatial multi omics in identifying precise therapeutic targets and predicting treatment responses while also highlighting the challenges that impede its integration into precision.
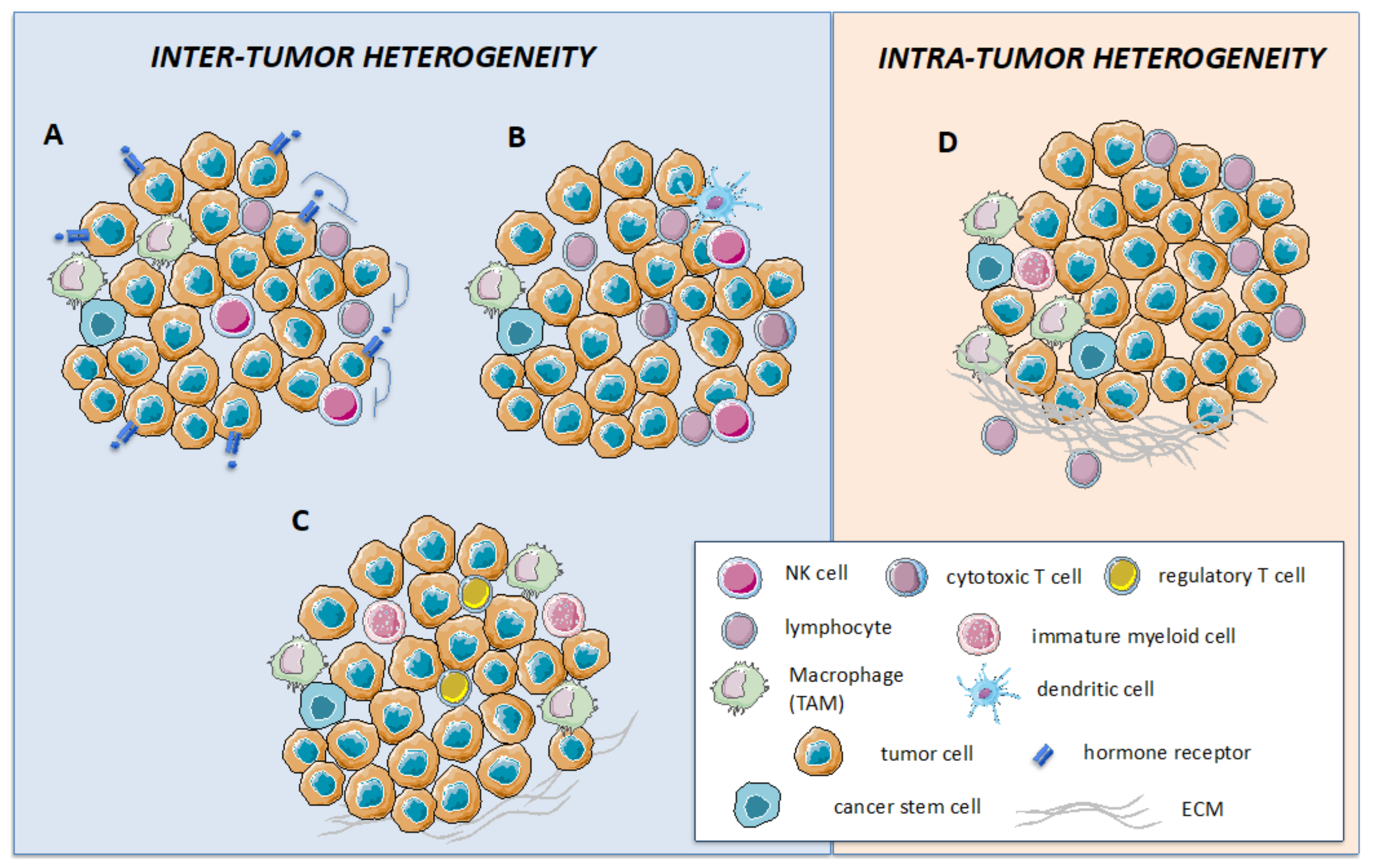
Cancers Free Full Text Impact Of Immune Cell Heterogeneity On He Background nasopharyngeal cancer (npc) is a squamous cell carcinoma of the upper pharynx, strongly associated with epstein barr virus (ebv), and with varying incidences in the world. we have previously investigated the presence and distribution of cd8 t cells in npc and found that an ‘ inflamed profile ’ , with cd8 t cells in and around cancer cell areas, was associated with better. Spatially resolved multi omics revolutionizes cancer therapy by decoding the cellular and molecular heterogeneity of the tumor microenvironment through spatial coordinates. this commentary discusses the roles of spatial multi omics in identifying precise therapeutic targets and predicting treatment responses while also highlighting the challenges that impede its integration into precision. Cell areas, was associated with better disease free survival (cf. non inflamed ‘desert’ tumours). in this study, we explore immune related biomarkers in a spatial context to assess distri bution of cell types and quantify selected biomarkers within the npc. methods using the geomx digital spatial profiler (dsp), 49. In a recent issue of nature, two back to back studies elegantly used 35 plex imc to characterize the single cell spatial landscapes of the brain [6] and lung [7] tme. by using specific markers to assign cellular lineage, cells in the tme were identified as ~20 components including cancer cells, endothelial cells, and immune cells (t cells, b.
Comments are closed.