Gene Ontology Enrichment Analysis Of Structural Reads From

Gene Ontology Enrichment Analysis Of Structural Reads From The number of genes in the interest gene set (ig.len), this represents the number of genes that are actually used for enrichment analysis. generally, a significant p value of the enrichment results should be less than 0.05 or 0.01, indicating a significant association between the interesting gene set and the disease node. Gene set enrichment analysis: a knowledge based approach for interpreting genome wide expression profiles. proceedings of the national academy of sciences 102 , 15545 lp–15550 (2005).
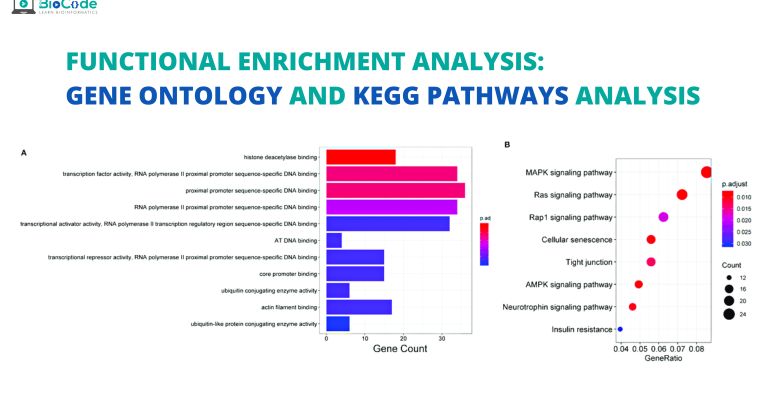
Enrichment Analysis Gene Ontology And Kegg Pathways Analysis Biocode Gene ontology enrichment analysis of structural reads from significantly changed proteins during 24 h macrophage activation. the chord plot presents the linkages of genes and go biological process. Gene ontology (go) enrichment analysis is ubiquitously used for interpreting high throughput molecular data and generating hypotheses about underlying biological phenomena of experiments. however. Functional enrichment analysis. to perform functional enrichment analysis, we need to have: a set of genes of interest (e.g., differentially expressed genes): study set. a set with all the genes to consider in the analysis: population set (which must contain the study set) go annotations, associating the genes in the population set to go terms. Go enrichment analysis. one of the main uses of the go is to perform enrichment analysis on gene sets. for example, given a set of genes that are up regulated under certain conditions, an enrichment analysis will find which go terms are over represented (or under represented) using annotations for that gene set.

Gene Ontology And Kegg Enrichment Analysis A Venn Diagram Showing Functional enrichment analysis. to perform functional enrichment analysis, we need to have: a set of genes of interest (e.g., differentially expressed genes): study set. a set with all the genes to consider in the analysis: population set (which must contain the study set) go annotations, associating the genes in the population set to go terms. Go enrichment analysis. one of the main uses of the go is to perform enrichment analysis on gene sets. for example, given a set of genes that are up regulated under certain conditions, an enrichment analysis will find which go terms are over represented (or under represented) using annotations for that gene set. Gene ontology enrichment analysis (goea) is used to test the overrepresentation of gene ontology terms in a list of genes or gene products in order to understand their biological significance. Background biological interpretation of gene protein lists resulting from omics experiments can be a complex task. a common approach consists of reviewing gene ontology (go) annotations for entries in such lists and searching for enrichment patterns. unfortunately, there is a gap between machine readable output of go software and its human interpretable form. this gap can be bridged by.

Gene Ontology Enrichment Analysis Of Structural Reads From Gene ontology enrichment analysis (goea) is used to test the overrepresentation of gene ontology terms in a list of genes or gene products in order to understand their biological significance. Background biological interpretation of gene protein lists resulting from omics experiments can be a complex task. a common approach consists of reviewing gene ontology (go) annotations for entries in such lists and searching for enrichment patterns. unfortunately, there is a gap between machine readable output of go software and its human interpretable form. this gap can be bridged by.
Comments are closed.