Single Cell Rna Sequencing Ngs Analysis

Single Cell Rna Sequencing Ngs Analysis Buettner, f. et al. computational analysis of cell to cell heterogeneity in single cell rna sequencing data reveals hidden subpopulations of cells. nat. biotechnol. 33 , 155–160 (2015). Single cell rna sequencing. scrna seq is a relatively new technology first introduced by tang et al. in 2009, but the cost of sequencing and limited number of protocols at the time meant that it did not get widespread popularity until 2014. scrnaseq differs from “traditional” bulk rnaseq. in bulk rnaseq, we measure the average expression of.

Single Cell Rna Sequencing Next Generation Sequencing Genewiz Rna sequencing (rna seq) is a genomic approach for the detection and quantitative analysis of messenger rna molecules in a biological sample and is useful for studying cellular responses. rna seq has fueled much discovery and innovation in medicine over recent years. for practical reasons, the technique is usually conducted on samples comprising thousands to millions of cells. however, this. 10x genomics’ single cell rna seq (scrna seq) technology, the chromium single cell 3’ solution, allows you to analyze transcriptomes on a cell by cell basis through the use of microfluidic partitioning to capture single cells and prepare barcoded, next generation sequencing (ngs) cdna libraries. Seurat is an r package designed for qc, analysis, and exploration of single cell rna seq data. developed and by the satija lab at the new york genome center. it is well maintained and well documented. it has a built in function to read 10x genomics data. it has implemented most of the steps needed in common analyses. Ngs based technologies for genomics, transcriptomics, and epigenomics are now increasingly focused on the characterization of individual cells. these single cell analyses will allow researchers to uncover new and potentially unexpected biological discoveries relative to traditional profiling methods that assess bulk populations. single cell rna.
Single Cell Rna Sequencing Ngs Analysis Seurat is an r package designed for qc, analysis, and exploration of single cell rna seq data. developed and by the satija lab at the new york genome center. it is well maintained and well documented. it has a built in function to read 10x genomics data. it has implemented most of the steps needed in common analyses. Ngs based technologies for genomics, transcriptomics, and epigenomics are now increasingly focused on the characterization of individual cells. these single cell analyses will allow researchers to uncover new and potentially unexpected biological discoveries relative to traditional profiling methods that assess bulk populations. single cell rna. As more analysis tools are becoming available, it is becoming increasingly difficult to navigate this landscape and produce an up to date workflow to analyse one's data. here, we detail the steps of a typical single cell rna seq analysis, including pre processing (quality control, normalization, data correction, feature selection, and. Next generation sequencing analysis resources. this e book contains resources for mastering ngs analysis. it has been generated by the bioinformatics team at nyu center for genomics and systems biology in new york and abu dhabi. the modules included in this resources are designed to provide hands on experience with analyzing next generation.
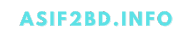
Comments are closed.